Difference between revisions of "Ontology"
Warner-admin (talk | contribs) m (Protected "Ontology" ([Edit=Allow only administrators] (indefinite) [Move=Allow only administrators] (indefinite))) |
Warner-admin (talk | contribs) |
||
| (19 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
| + | <span id="BackToTop"></span> | ||
| + | <div class="noprint" style="background-color:LightGray; position:fixed; bottom:2%; right:0.25%; padding-left:5px; padding-right:5px; margin: 15px; opacity:0.8; border-style: solid; border-color:DarkGray; border-width: 1px"> | ||
| + | [[#top|Back to Top]] | ||
| + | </div> | ||
| + | Note that the structure and content of HemOnc.org are constantly undergoing evolution, such that information that you read here might be outdated. This page was last reviewed and updated in '''May 2023'''. | ||
| + | |||
==Introduction to HemOnc vocabulary== | ==Introduction to HemOnc vocabulary== | ||
| − | In late 2018, we began to work closely with the [https://www.ohdsi.org/ OHDSI Oncology Subgroup] to transition from the existing OWL format to the widely-used OMOP format. The first phase of this work was completed in June 2019 and a portion of the HemOnc vocabulary has been released to the public through the [http://athena.ohdsi.org/search-terms/terms ATHENA] tool.You can see how this looks by searching for "GemOx" using [http://athena.ohdsi.org/search-terms/terms?query=gemox&page=1 this link | + | In late 2018, we began to work closely with the [https://www.ohdsi.org/ OHDSI Oncology Subgroup] to transition from the existing [[#HemOnc_ontology_in_OWL_format_.28historical.29|OWL format]] to the widely-used OMOP format. The first phase of this work was completed in June 2019 and a portion of the HemOnc vocabulary has been released to the public through the [http://athena.ohdsi.org/search-terms/terms ATHENA] tool.You can see how this looks by searching for "GemOx" using [http://athena.ohdsi.org/search-terms/terms?query=gemox&page=1 this link]. |
The OMOP model is built on [[Vocabulary concepts|concept]] and [[Vocabulary relationships|relationship]] tables. Please use the links to learn more about what concepts and relationships are currently included in the vocabulary. | The OMOP model is built on [[Vocabulary concepts|concept]] and [[Vocabulary relationships|relationship]] tables. Please use the links to learn more about what concepts and relationships are currently included in the vocabulary. | ||
==Availability== | ==Availability== | ||
| − | Beyond what is available through [http://athena.ohdsi.org/search-terms/terms ATHENA], the full ontology is freely available for academic or non-commercial use | + | Beyond what is available through [http://athena.ohdsi.org/search-terms/terms ATHENA], the full ontology is freely available for academic or non-commercial use. A subset is available to any user, see below. Both versions can be accessed through the [https://dataverse.harvard.edu/dataverse/HemOnc HemOnc Dataverse]. |
| − | ==Academic and Non- | + | We are also willing to discuss commercial uses on a case-by-case basis. Please let us know of your interest by [mailto:[email protected] sending an email] (<u>''[email protected]</u>''). |
| − | The full HemOnc | + | |
| + | ==Full Academic and Non-Commercial License== | ||
| + | The [https://dataverse.harvard.edu/dataset.xhtml?persistentId=doi:10.7910/DVN/FPO4HB full HemOnc ontology] is licensed by version under the [https://creativecommons.org/licenses/by-nc-sa/4.0/ '''CreativeCommons NonCommercial-ShareAlike 4.0 International License (CC BY-NC-SA 4.0)'''] for academic and non-commercial users. Under this license, users are free to: | ||
*'''Share''' — copy and redistribute the material in any medium or format | *'''Share''' — copy and redistribute the material in any medium or format | ||
| Line 19: | Line 27: | ||
*'''ShareAlike''' — If you remix, transform, or build upon the material, you must distribute your contributions under the same license as the original. | *'''ShareAlike''' — If you remix, transform, or build upon the material, you must distribute your contributions under the same license as the original. | ||
*'''No additional restrictions''' — You may not apply legal terms or technological measures that legally restrict others from doing anything the license permits. | *'''No additional restrictions''' — You may not apply legal terms or technological measures that legally restrict others from doing anything the license permits. | ||
| + | |||
| + | ==Subset License== | ||
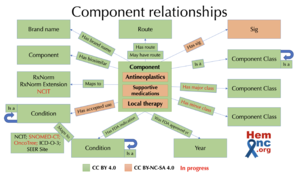
| + | A [https://dataverse.harvard.edu/dataset.xhtml?persistentId=doi:10.7910/DVN/9CY9C6 subset of HemOnc] focused on components and their relationships is licensed under the [https://creativecommons.org/licenses/by/4.0/ '''CreativeCommons Attribution 4.0 International License (CC BY 4.0)'''] for any user. | ||
| + | |||
| + | [[File:HemOnc component rels.png|thumb]] | ||
| + | |||
| + | Under this license, users are free to: | ||
| + | |||
| + | *'''Share''' — copy and redistribute the material in any medium or format | ||
| + | *'''Adapt''' — remix, transform, and build upon the material for any purpose, even commercially. | ||
| + | |||
| + | Under the following terms: | ||
| + | |||
| + | *'''Attribution''' — You must give appropriate credit, provide a link to the license, and indicate if changes were made. You may do so in any reasonable manner, but not in any way that suggests the licensor endorses you or your use. | ||
| + | *'''No additional restrictions''' — You may not apply legal terms or technological measures that legally restrict others from doing anything the license permits. | ||
| + | |||
| + | ==FAQ== | ||
| + | The purpose of this section is to address some common questions on how to pull out subsets of the data; it will be expanded over time. For simple cross-tabulations, please visit our data browser, accessible via the link in the banner. The answers below are provided using R code. | ||
| + | ===Q: How would you query the ontology to find all Immune Checkpoint Inhibitors?=== | ||
| + | '''A:''' Immune checkpoint inhibition is one of the most important advances in oncology in the 2010s and beyond. The first commercial products targeted CTLA-4, PD-1, and PD-L1; however, there are [[:Category:Immunostimulatory_monoclonal_antibodies|many additional immunostimulatory targets]]. Therefore, we preferentially group drugs by their specific mechanism of action, such as anti-CTLA-4 antibodies, anti-PD-1 antibodies, and so forth. If one wanted a list of all of the anti-CTLA-4 antibodies, anti-PD-1 antibodies, and anti-PD-L1 antibodies, the following query would work: | ||
| + | |||
| + | targets <- concept_stage$concept_code[concept_stage$concept_name %in% c('Anti-CTLA-4 antibody','Anti-PD-1 antibody','Anti-PD-L1 antibody')] | ||
| + | temp.ref <- which(concept_relationship_stage$concept_code_2 %in% targets & concept_relationship_stage$relationship_id == 'Is a') | ||
| + | result <- concept_stage[concept_stage$concept_code %in% concept_relationship_stage$concept_code_1[temp.ref],] | ||
| + | |||
| + | If one wanted the full list of immunostimulatory monoclonal antibodies, one could use the following query: | ||
| + | |||
| + | target <- concept_stage$concept_code[concept_stage$concept_name %in% c('Immunostimulatory monoclonal antibody')] | ||
| + | temp.ref <- which(concept_relationship_stage$concept_code_2 %in% target & concept_relationship_stage$relationship_id == 'Is a') | ||
| + | temp.ref <- which(concept_relationship_stage$concept_code_2 %in% concept_relationship_stage$concept_code_1[temp.ref] & concept_relationship_stage$relationship_id == 'Is a') | ||
| + | result <- concept_stage[concept_stage$concept_code %in% concept_relationship_stage$concept_code_1[temp.ref],] | ||
| + | |||
| + | Finally, if one were only interested in FDA-approved anti-CTLA-4 antibodies, anti-PD-1 antibodies, and anti-PD-L1 antibodies, the following query would work: | ||
| + | |||
| + | targets <- concept_stage$concept_code[concept_stage$concept_name %in% c('Anti-CTLA-4 antibody','Anti-PD-1 antibody','Anti-PD-L1 antibody')] | ||
| + | temp.ref <- which(concept_relationship_stage$concept_code_2 %in% targets & concept_relationship_stage$relationship_id == 'Is a') | ||
| + | temp.ref <- which(concept_relationship_stage$concept_code_1 %in% concept_relationship_stage$concept_code_1[temp.ref] & concept_relationship_stage$relationship_id == 'Was FDA approved yr') | ||
| + | result <- concept_stage[concept_stage$concept_code %in% concept_relationship_stage$concept_code_1[temp.ref],] | ||
==References and acknowledgements== | ==References and acknowledgements== | ||
| − | # Malty AM, Jain SK, Yang PC, Harvey K, Warner JL. Computerized approach to creating a systematic ontology of hematology/oncology regimens. JCO Clin Cancer Inform. 2018 May 11. [https://doi.org/10.1200/CCI.17.00142 link to original article] [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6141041/ link to PMC article] [https://pubmed.ncbi.nlm.nih.gov/30238070 PubMed] | + | # Malty AM, Jain SK, Yang PC, Harvey K, Warner JL. Computerized approach to creating a systematic ontology of hematology/oncology regimens. JCO Clin Cancer Inform. 2018 May 11. [https://doi.org/10.1200/CCI.17.00142 link to original article] [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6141041/ link to PMC article] [https://pubmed.ncbi.nlm.nih.gov/30238070/ PubMed] |
| − | # Warner JL, Dymshyts D, Reich CG, Gurley MJ, Hochheiser H, Moldwin ZH, Belenkaya R, Williams AE, Yang PC. HemOnc: a new standard vocabulary for chemotherapy regimen representation in the OMOP Common Data Model. J Biomed Inform. 2019 Aug;96:103239. Epub 2019 Jun 22. [https://www. | + | # Warner JL, Dymshyts D, Reich CG, Gurley MJ, Hochheiser H, Moldwin ZH, Belenkaya R, Williams AE, Yang PC. HemOnc: a new standard vocabulary for chemotherapy regimen representation in the OMOP Common Data Model. J Biomed Inform. 2019 Aug;96:103239. Epub 2019 Jun 22. [https://doi.org/10.1016/j.jbi.2019.103239 link to original article] [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6697579/ link to PMC article] [https://pubmed.ncbi.nlm.nih.gov/31238109/ PubMed] |
| − | # This work is currently supported in part by National Institutes of Health (NIH) Grants No. [ | + | # Rubinstein SM, Yang PC, Cowan AJ, Warner JL. Standardizing Chemotherapy Regimen Nomenclature: A Proposal and Evaluation of the HemOnc and National Cancer Institute Thesaurus Regimen Content. JCO Clin Cancer Inform. 2020 Jan;4:60-70. [https://doi.org/10.1200/cci.19.00122 link to original article] [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7000232/ link to PMC article] [https://pubmed.ncbi.nlm.nih.gov/31990580/ PubMed] |
| + | # Rubinstein SM, Sethi T, Venepalli NK, Gyawali B, Schwartz C, Rivera DR, Yang PC, Warner JL. Chemotherapy Knowledge Base Management in the Era of Precision Oncology. JCO Clin Cancer Inform. 2021 Jan;5:30-35. [https://doi.org/10.1200/cci.20.00076 link to original article] [https://pubmed.ncbi.nlm.nih.gov/33411619/ PubMed] | ||
| + | # This work is currently supported in part by National Institutes of Health (NIH) Grants No. [https://grantome.com/grant/NIH/U24-CA248010-01A1 U24 CA248010] and [https://reporter.nih.gov/search/bweQKIeIwk-kOtL_oHSGSw/project-details/10705015 U24 CA265879]. Previously, this work was supported by NIH Grant No. L30 CA171123, [http://grantome.com/grant/NIH/U24-CA184407-01 U24 CA184407], [https://grantome.com/grant/NIH/U24-CA194215-01A1 U24 CA194215], and [https://grantome.com/grant/NIH/U01-CA231840-01 U01 CA231840]. Portions of this work were conducted using the [https://protege.stanford.edu/ Protégé resource], which is supported by Grant No. GM10331601 from the NIH National Institute of General Medical Sciences. | ||
| − | == | + | ==HemOnc ontology in OWL format (historical)== |
| − | In the summer of 2017, we began the process of converting portions of the HemOnc.org website content into a formal [https://www.w3.org/OWL/ OWL ontology], along with some additional external information such as RxNorm identifiers. As of the last release ('''2018-10-29'''), the ontology | + | In the summer of 2017, we began the process of converting portions of the HemOnc.org website content into a formal [https://www.w3.org/OWL/ OWL ontology], along with some additional external information such as RxNorm identifiers. As of the last release ('''2018-10-29'''), the ontology contained information on: |
*'''Diseases:''' names, classifications | *'''Diseases:''' names, classifications | ||
| Line 40: | Line 88: | ||
For a much more detailed explanation, you can download the latest release notes here: [[File:Release notes 2018-10-29.docx]]. | For a much more detailed explanation, you can download the latest release notes here: [[File:Release notes 2018-10-29.docx]]. | ||
| − | <span style="color:red">'''Note: due to a shift to an OMOP-compatible database, we do not plan to support additional versions of the ontology in OWL, for the time being | + | <span style="color:red">'''Note: due to a shift to an OMOP-compatible database, we do not plan to support additional versions of the ontology in OWL, for the time being.'''</span> |
| − | [[Category: | + | [[Category:Ontology pages]] |
Latest revision as of 14:24, 30 December 2023
Note that the structure and content of HemOnc.org are constantly undergoing evolution, such that information that you read here might be outdated. This page was last reviewed and updated in May 2023.
Introduction to HemOnc vocabulary
In late 2018, we began to work closely with the OHDSI Oncology Subgroup to transition from the existing OWL format to the widely-used OMOP format. The first phase of this work was completed in June 2019 and a portion of the HemOnc vocabulary has been released to the public through the ATHENA tool.You can see how this looks by searching for "GemOx" using this link.
The OMOP model is built on concept and relationship tables. Please use the links to learn more about what concepts and relationships are currently included in the vocabulary.
Availability
Beyond what is available through ATHENA, the full ontology is freely available for academic or non-commercial use. A subset is available to any user, see below. Both versions can be accessed through the HemOnc Dataverse.
We are also willing to discuss commercial uses on a case-by-case basis. Please let us know of your interest by sending an email ([email protected]).
Full Academic and Non-Commercial License
The full HemOnc ontology is licensed by version under the CreativeCommons NonCommercial-ShareAlike 4.0 International License (CC BY-NC-SA 4.0) for academic and non-commercial users. Under this license, users are free to:
- Share — copy and redistribute the material in any medium or format
- Adapt — remix, transform, and build upon the material
Under the following terms:
- Attribution — You must give appropriate credit, provide a link to the license, and indicate if changes were made. You may do so in any reasonable manner, but not in any way that suggests the licensor endorses you or your use.
- NonCommercial — You may not use the material for commercial purposes.
- ShareAlike — If you remix, transform, or build upon the material, you must distribute your contributions under the same license as the original.
- No additional restrictions — You may not apply legal terms or technological measures that legally restrict others from doing anything the license permits.
Subset License
A subset of HemOnc focused on components and their relationships is licensed under the CreativeCommons Attribution 4.0 International License (CC BY 4.0) for any user.
Under this license, users are free to:
- Share — copy and redistribute the material in any medium or format
- Adapt — remix, transform, and build upon the material for any purpose, even commercially.
Under the following terms:
- Attribution — You must give appropriate credit, provide a link to the license, and indicate if changes were made. You may do so in any reasonable manner, but not in any way that suggests the licensor endorses you or your use.
- No additional restrictions — You may not apply legal terms or technological measures that legally restrict others from doing anything the license permits.
FAQ
The purpose of this section is to address some common questions on how to pull out subsets of the data; it will be expanded over time. For simple cross-tabulations, please visit our data browser, accessible via the link in the banner. The answers below are provided using R code.
Q: How would you query the ontology to find all Immune Checkpoint Inhibitors?
A: Immune checkpoint inhibition is one of the most important advances in oncology in the 2010s and beyond. The first commercial products targeted CTLA-4, PD-1, and PD-L1; however, there are many additional immunostimulatory targets. Therefore, we preferentially group drugs by their specific mechanism of action, such as anti-CTLA-4 antibodies, anti-PD-1 antibodies, and so forth. If one wanted a list of all of the anti-CTLA-4 antibodies, anti-PD-1 antibodies, and anti-PD-L1 antibodies, the following query would work:
targets <- concept_stage$concept_code[concept_stage$concept_name %in% c('Anti-CTLA-4 antibody','Anti-PD-1 antibody','Anti-PD-L1 antibody')]
temp.ref <- which(concept_relationship_stage$concept_code_2 %in% targets & concept_relationship_stage$relationship_id == 'Is a')
result <- concept_stage[concept_stage$concept_code %in% concept_relationship_stage$concept_code_1[temp.ref],]
If one wanted the full list of immunostimulatory monoclonal antibodies, one could use the following query:
target <- concept_stage$concept_code[concept_stage$concept_name %in% c('Immunostimulatory monoclonal antibody')]
temp.ref <- which(concept_relationship_stage$concept_code_2 %in% target & concept_relationship_stage$relationship_id == 'Is a')
temp.ref <- which(concept_relationship_stage$concept_code_2 %in% concept_relationship_stage$concept_code_1[temp.ref] & concept_relationship_stage$relationship_id == 'Is a')
result <- concept_stage[concept_stage$concept_code %in% concept_relationship_stage$concept_code_1[temp.ref],]
Finally, if one were only interested in FDA-approved anti-CTLA-4 antibodies, anti-PD-1 antibodies, and anti-PD-L1 antibodies, the following query would work:
targets <- concept_stage$concept_code[concept_stage$concept_name %in% c('Anti-CTLA-4 antibody','Anti-PD-1 antibody','Anti-PD-L1 antibody')]
temp.ref <- which(concept_relationship_stage$concept_code_2 %in% targets & concept_relationship_stage$relationship_id == 'Is a')
temp.ref <- which(concept_relationship_stage$concept_code_1 %in% concept_relationship_stage$concept_code_1[temp.ref] & concept_relationship_stage$relationship_id == 'Was FDA approved yr')
result <- concept_stage[concept_stage$concept_code %in% concept_relationship_stage$concept_code_1[temp.ref],]
References and acknowledgements
- Malty AM, Jain SK, Yang PC, Harvey K, Warner JL. Computerized approach to creating a systematic ontology of hematology/oncology regimens. JCO Clin Cancer Inform. 2018 May 11. link to original article link to PMC article PubMed
- Warner JL, Dymshyts D, Reich CG, Gurley MJ, Hochheiser H, Moldwin ZH, Belenkaya R, Williams AE, Yang PC. HemOnc: a new standard vocabulary for chemotherapy regimen representation in the OMOP Common Data Model. J Biomed Inform. 2019 Aug;96:103239. Epub 2019 Jun 22. link to original article link to PMC article PubMed
- Rubinstein SM, Yang PC, Cowan AJ, Warner JL. Standardizing Chemotherapy Regimen Nomenclature: A Proposal and Evaluation of the HemOnc and National Cancer Institute Thesaurus Regimen Content. JCO Clin Cancer Inform. 2020 Jan;4:60-70. link to original article link to PMC article PubMed
- Rubinstein SM, Sethi T, Venepalli NK, Gyawali B, Schwartz C, Rivera DR, Yang PC, Warner JL. Chemotherapy Knowledge Base Management in the Era of Precision Oncology. JCO Clin Cancer Inform. 2021 Jan;5:30-35. link to original article PubMed
- This work is currently supported in part by National Institutes of Health (NIH) Grants No. U24 CA248010 and U24 CA265879. Previously, this work was supported by NIH Grant No. L30 CA171123, U24 CA184407, U24 CA194215, and U01 CA231840. Portions of this work were conducted using the Protégé resource, which is supported by Grant No. GM10331601 from the NIH National Institute of General Medical Sciences.
HemOnc ontology in OWL format (historical)
In the summer of 2017, we began the process of converting portions of the HemOnc.org website content into a formal OWL ontology, along with some additional external information such as RxNorm identifiers. As of the last release (2018-10-29), the ontology contained information on:
- Diseases: names, classifications
- Interventions, in four major classes:
- Biologic products: factor replacement, cellular therapy, etc.
- Devices
- Drugs: names, synonyms, classifications, RxCUI (when available), sigs
- Radiation
- Regimens: names, synonyms, classifications, components, references
- References: title, journal, year of publication, classifications, number of authors, authors
- Authors: name
For a much more detailed explanation, you can download the latest release notes here: File:Release notes 2018-10-29.docx.
Note: due to a shift to an OMOP-compatible database, we do not plan to support additional versions of the ontology in OWL, for the time being.